Easily extract useful features from character objects.
Usage
textfeatures()
Input a character vector.
## vector of some text
x <- c(
"this is A!\t sEntence https://github.com about #rstats @github",
"and another sentence here", "THe following list:\n- one\n- two\n- three\nOkay!?!"
)
## get text features
textfeatures(x, verbose = FALSE)
#> # A tibble: 3 x 36
#> n_urls n_uq_urls n_hashtags n_uq_hashtags n_mentions n_uq_mentions n_chars n_uq_chars n_commas
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1.15 1.15 1.15 1.15 1.15 1.15 0.243 0.330 0
#> 2 -0.577 -0.577 -0.577 -0.577 -0.577 -0.577 -1.10 -1.12 0
#> 3 -0.577 -0.577 -0.577 -0.577 -0.577 -0.577 0.856 0.793 0
#> # … with 27 more variables: n_digits <dbl>, n_exclaims <dbl>, n_extraspaces <dbl>, n_lowers <dbl>,
#> # n_lowersp <dbl>, n_periods <dbl>, n_words <dbl>, n_uq_words <dbl>, n_caps <dbl>,
#> # n_nonasciis <dbl>, n_puncts <dbl>, n_capsp <dbl>, n_charsperword <dbl>, sent_afinn <dbl>,
#> # sent_bing <dbl>, sent_syuzhet <dbl>, sent_vader <dbl>, n_polite <dbl>, n_first_person <dbl>,
#> # n_first_personp <dbl>, n_second_person <dbl>, n_second_personp <dbl>, n_third_person <dbl>,
#> # n_tobe <dbl>, n_prepositions <dbl>, w1 <dbl>, w2 <dbl>Or input a data frame with a column named text.
## data frame with rstats tweets
rt <- rtweet::search_tweets("rstats", n = 2000, verbose = FALSE)
## get text features
tf <- textfeatures(rt, verbose = FALSE)
## preview data
tf
#> # A tibble: 2,000 x 134
#> n_urls n_uq_urls n_hashtags n_uq_hashtags n_mentions n_uq_mentions n_chars n_uq_chars n_commas
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0.838 0.336 1.34 1.34 -0.564 -0.564 0.770 0.749 -0.605
#> 2 -0.338 0.336 -0.636 -0.634 -0.564 -0.564 -0.815 -0.958 -0.605
#> 3 -2.35 -2.92 -1.06 -1.06 3.70 3.70 0.498 0.903 -0.605
#> 4 -0.338 0.336 -0.636 -0.634 -0.564 -0.564 -0.561 -0.675 -0.605
#> 5 0.838 0.336 0.385 0.386 1.09 1.09 -0.773 -0.813 -0.605
#> 6 -0.338 0.336 -1.06 -1.06 1.09 1.09 0.0764 -0.420 2.35
#> 7 -0.338 0.336 -0.636 -0.634 2.05 2.05 -0.195 -0.0763 -0.605
#> 8 -2.35 -2.92 -1.06 -1.06 1.09 1.09 -0.181 0.130 1.74
#> 9 -0.338 0.336 -1.06 -1.06 3.27 3.27 0.406 0.827 1.74
#> 10 -2.35 -2.92 -0.636 -0.634 -0.564 -0.564 -0.283 -0.300 -0.605
#> # … with 1,990 more rows, and 125 more variables: n_digits <dbl>, n_exclaims <dbl>,
#> # n_extraspaces <dbl>, n_lowers <dbl>, n_lowersp <dbl>, n_periods <dbl>, n_words <dbl>,
#> # n_uq_words <dbl>, n_caps <dbl>, n_nonasciis <dbl>, n_puncts <dbl>, n_capsp <dbl>,
#> # n_charsperword <dbl>, sent_afinn <dbl>, sent_bing <dbl>, sent_syuzhet <dbl>, sent_vader <dbl>,
#> # n_polite <dbl>, n_first_person <dbl>, n_first_personp <dbl>, n_second_person <dbl>,
#> # n_second_personp <dbl>, n_third_person <dbl>, n_tobe <dbl>, n_prepositions <dbl>, w1 <dbl>,
#> # w2 <dbl>, w3 <dbl>, w4 <dbl>, w5 <dbl>, w6 <dbl>, w7 <dbl>, w8 <dbl>, w9 <dbl>, w10 <dbl>,
#> # w11 <dbl>, w12 <dbl>, w13 <dbl>, w14 <dbl>, w15 <dbl>, w16 <dbl>, w17 <dbl>, w18 <dbl>,
#> # w19 <dbl>, w20 <dbl>, w21 <dbl>, w22 <dbl>, w23 <dbl>, w24 <dbl>, w25 <dbl>, w26 <dbl>,
#> # w27 <dbl>, w28 <dbl>, w29 <dbl>, w30 <dbl>, w31 <dbl>, w32 <dbl>, w33 <dbl>, w34 <dbl>,
#> # w35 <dbl>, w36 <dbl>, w37 <dbl>, w38 <dbl>, w39 <dbl>, w40 <dbl>, w41 <dbl>, w42 <dbl>,
#> # w43 <dbl>, w44 <dbl>, w45 <dbl>, w46 <dbl>, w47 <dbl>, w48 <dbl>, w49 <dbl>, w50 <dbl>,
#> # w51 <dbl>, w52 <dbl>, w53 <dbl>, w54 <dbl>, w55 <dbl>, w56 <dbl>, w57 <dbl>, w58 <dbl>,
#> # w59 <dbl>, w60 <dbl>, w61 <dbl>, w62 <dbl>, w63 <dbl>, w64 <dbl>, w65 <dbl>, w66 <dbl>,
#> # w67 <dbl>, w68 <dbl>, w69 <dbl>, w70 <dbl>, w71 <dbl>, w72 <dbl>, w73 <dbl>, w74 <dbl>,
#> # w75 <dbl>, …Compare across multiple authors.
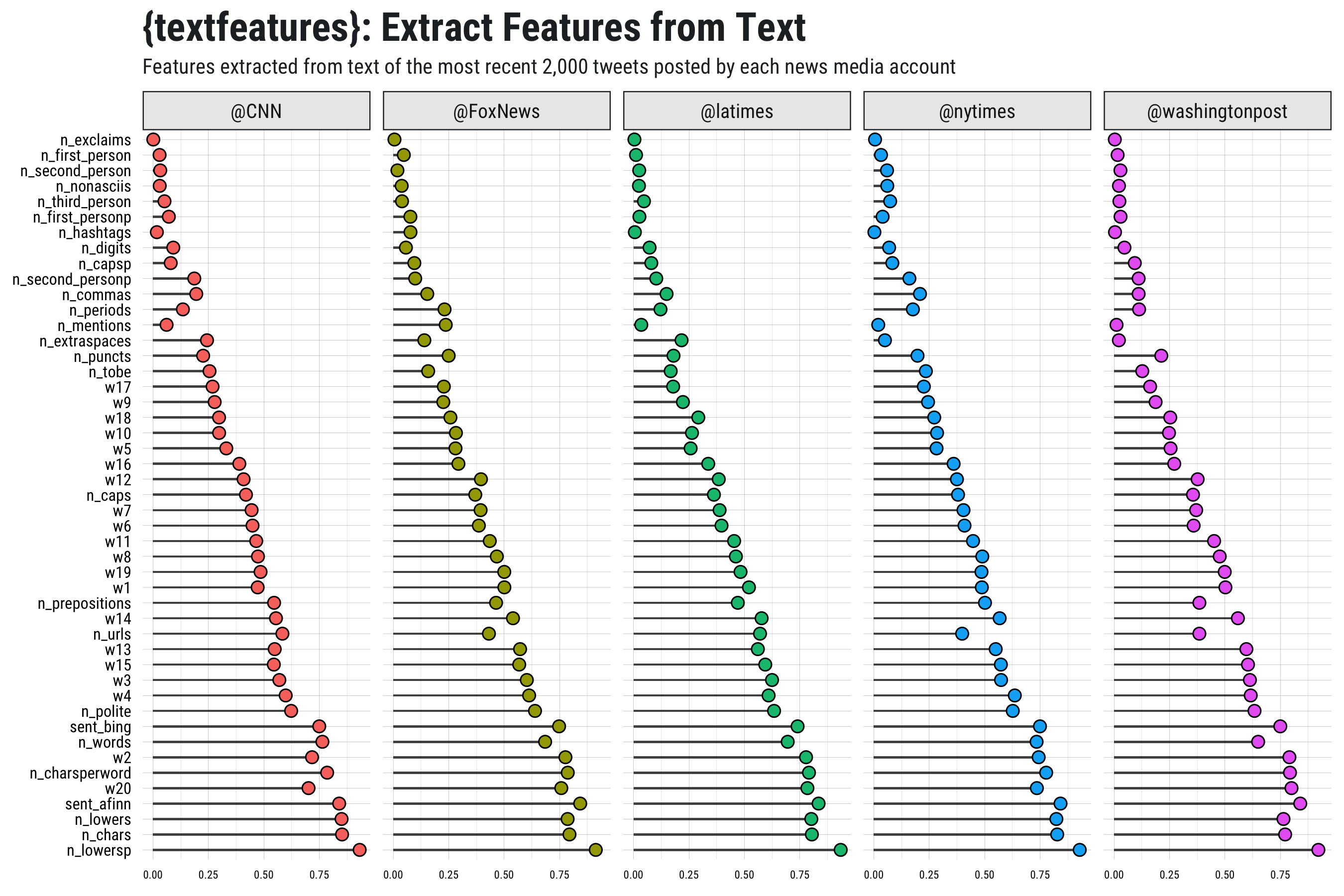
## data frame tweets from multiple news media accounts
news <- rtweet::get_timelines(
c("cnn", "nytimes", "foxnews", "latimes", "washingtonpost"),
n = 2000)
## get text features (including ests for 20 word dims) for all observations
news_features <- textfeatures(news, word_dims = 20, verbose = FALSE)
Fast version
If you’re looking for something faster try setting sentiment = FALSE and word2vec = 0.
## get non-substantive text features
textfeatures(rt, sentiment = FALSE, word_dims = 0, verbose = FALSE)
#> # A tibble: 2,000 x 29
#> n_urls n_uq_urls n_hashtags n_uq_hashtags n_mentions n_uq_mentions n_chars n_uq_chars n_commas
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 0.838 0.336 1.34 1.34 -0.564 -0.564 0.770 0.749 -0.605
#> 2 -0.338 0.336 -0.636 -0.634 -0.564 -0.564 -0.815 -0.958 -0.605
#> 3 -2.35 -2.92 -1.06 -1.06 3.70 3.70 0.498 0.903 -0.605
#> 4 -0.338 0.336 -0.636 -0.634 -0.564 -0.564 -0.561 -0.675 -0.605
#> 5 0.838 0.336 0.385 0.386 1.09 1.09 -0.773 -0.813 -0.605
#> 6 -0.338 0.336 -1.06 -1.06 1.09 1.09 0.0764 -0.420 2.35
#> 7 -0.338 0.336 -0.636 -0.634 2.05 2.05 -0.195 -0.0763 -0.605
#> 8 -2.35 -2.92 -1.06 -1.06 1.09 1.09 -0.181 0.130 1.74
#> 9 -0.338 0.336 -1.06 -1.06 3.27 3.27 0.406 0.827 1.74
#> 10 -2.35 -2.92 -0.636 -0.634 -0.564 -0.564 -0.283 -0.300 -0.605
#> # … with 1,990 more rows, and 20 more variables: n_digits <dbl>, n_exclaims <dbl>,
#> # n_extraspaces <dbl>, n_lowers <dbl>, n_lowersp <dbl>, n_periods <dbl>, n_words <dbl>,
#> # n_uq_words <dbl>, n_caps <dbl>, n_nonasciis <dbl>, n_puncts <dbl>, n_capsp <dbl>,
#> # n_charsperword <dbl>, n_first_person <dbl>, n_first_personp <dbl>, n_second_person <dbl>,
#> # n_second_personp <dbl>, n_third_person <dbl>, n_tobe <dbl>, n_prepositions <dbl>Example: NASA meta data
Extract text features from NASA meta data:
## read NASA meta data
nasa <- jsonlite::fromJSON("https://data.nasa.gov/data.json")
## identify non-public or restricted data sets
nonpub <- grepl("Not publicly available|must register",
nasa$data$rights, ignore.case = TRUE) |
nasa$dataset$accessLevel %in% c("restricted public", "non-public")
## create data frame with ID, description (name it "text"), and nonpub
nd <- data.frame(text = nasa$dataset$description, nonpub = nonpub,
stringsAsFactors = FALSE)
## drop duplicates (truncate text to ensure more distinct obs)
nd <- nd[!duplicated(tolower(substr(nd$text, 1, 100))), ]
## filter via sampling to create equal number of pub/nonpub
nd <- nd[c(sample(which(!nd$nonpub), sum(nd$nonpub)), which(nd$nonpub)), ]## get text features
nasa_tf <- textfeatures(nd, word_dims = 20, normalize = FALSE, verbose = FALSE)
## drop columns with little to no variance
min_var <- function(x, min = 1) {
is_num <- vapply(x, is.numeric, logical(1))
non_num <- names(x)[!is_num]
yminvar <- names(x[is_num])[vapply(x[is_num], function(.x) stats::var(.x,
na.rm = TRUE) >= min, logical(1))]
x[c(non_num, yminvar)]
}
nasa_tf <- min_var(nasa_tf)
## view summary
skimrskim(nasa_tf)| variable | min | 25% | mid | 75% | max | hist |
|---|---|---|---|---|---|---|
| n_caps | 1 | 10 | 28 | 46 | 207 | ▇▇▂▁▁▁▁▁ |
| n_commas | 0 | 1 | 6 | 9.75 | 32 | ▇▅▃▁▁▁▁▁ |
| n_digits | 0 | 0 | 2 | 6 | 57 | ▇▁▁▁▁▁▁▁ |
| n_extraspaces | 0 | 0 | 0 | 0 | 29 | ▇▁▁▁▁▁▁▁ |
| n_lowers | 0 | 4.25 | 47 | 853.5 | 3123 | ▇▁▂▁▁▁▁▁ |
| n_nonasciis | 0 | 0 | 0 | 0 | 20 | ▇▁▁▁▁▁▁▁ |
| n_periods | 0 | 0 | 2 | 6 | 28 | ▇▂▁▁▁▁▁▁ |
| n_prepositions | 0 | 0 | 1 | 8 | 18 | ▇▁▁▃▂▁▁▁ |
| n_puncts | 0 | 0 | 2 | 12 | 59 | ▇▂▁▁▁▁▁▁ |
| n_tobe | 0 | 0 | 0 | 3 | 7 | ▇▁▁▂▁▁▁▁ |
| n_uq_chars | 2 | 15 | 28.5 | 46 | 68 | ▂▇▅▂▅▅▃▁ |
| n_uq_words | 1 | 7 | 12.5 | 112.75 | 341 | ▇▂▂▂▁▁▁▁ |
| n_words | 1 | 7 | 12.5 | 163.5 | 598 | ▇▂▂▁▁▁▁▁ |
| sent_afinn | -18 | 0 | 0 | 3 | 30 | ▁▁▇▂▁▁▁▁ |
| sent_bing | -9 | 0 | 0 | 1 | 23 | ▁▁▇▁▁▁▁▁ |
| sent_syuzhet | -3.5 | 0 | 0 | 4.16 | 32.25 | ▇▂▂▁▁▁▁▁ |
| sent_vader | -11.5 | 0 | 0 | 2.8 | 31.4 | ▁▁▇▁▁▁▁▁ |
## add nonpub variable
nasa_tf$nonpub <- nd$nonpub
## run model predicting whether data is restricted
m1 <- glm(nonpub ~ ., data = nasa_tf[-1], family = binomial)
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
## view model summary
summary(m1)
#>
#> Call:
#> glm(formula = nonpub ~ ., family = binomial, data = nasa_tf[-1])
#>
#> Deviance Residuals:
#> Min 1Q Median 3Q Max
#> -2.01381 -0.01885 0.00078 0.04314 2.29757
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 8.31318 2.70503 3.073 0.00212 **
#> n_uq_chars -0.37317 0.14005 -2.665 0.00771 **
#> n_commas 0.14884 0.25324 0.588 0.55671
#> n_digits -0.19962 0.13118 -1.522 0.12809
#> n_extraspaces 0.08942 0.16235 0.551 0.58179
#> n_lowers -0.01618 0.03261 -0.496 0.61983
#> n_periods 1.17591 0.44971 2.615 0.00893 **
#> n_words -0.02638 0.14660 -0.180 0.85723
#> n_uq_words 0.04423 0.17763 0.249 0.80337
#> n_caps 0.17170 0.06327 2.714 0.00666 **
#> n_nonasciis -1.77660 367.21424 -0.005 0.99614
#> n_puncts -0.21932 0.16775 -1.307 0.19107
#> sent_afinn 0.19473 0.43352 0.449 0.65330
#> sent_bing -0.56450 0.56620 -0.997 0.31876
#> sent_syuzhet 0.06075 0.59648 0.102 0.91888
#> sent_vader -0.09451 0.35599 -0.265 0.79064
#> n_tobe -0.49601 0.76199 -0.651 0.51509
#> n_prepositions 0.21984 0.52947 0.415 0.67799
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 396.480 on 285 degrees of freedom
#> Residual deviance: 57.512 on 268 degrees of freedom
#> AIC: 93.512
#>
#> Number of Fisher Scoring iterations: 19
## how accurate was the model?
table(predict(m1, type = "response") > .5, nasa_tf$nonpub)
#>
#> FALSE TRUE
#> FALSE 138 7
#> TRUE 5 136
